So how is this construct created? The first step is to identify the sequence of the promoter region of F45D3.4 (where the transcription factors bind). Once identified, primers can be used in order to clone and amplify the promoter region. Primers are short strands of artificial DNA that work by being a complementary pair to the DNA strand of interest. Once the primer binds to the promoter of F45D3.4, DNA-polymerase can bind and can synthesize this new double stranded DNA, thus amplifying the amount of the promoter region (a detailed explanation of how primers work can be viewed in the support page of qPCR) [1].
Padmanabha D, Padilla PA, You Y, Baker KD (2015)
A HIF-Independent Mediator of Transcriptional Responses to Oxygen
by Deprivation in Caenorhabditis Elegans
Genetics, Vol. 199, 739-748
A HIF-Independent Mediator of Transcriptional Responses to Oxygen
by Deprivation in Caenorhabditis Elegans
Genetics, Vol. 199, 739-748
(Translated by Emaan Chaudry)
Visualizing genes with GFP under the microscope:
Microscopy was used on GFP-expressing worms using a light microscope. But how exactly were the researchers able to get the worms to light up? Green fluorescent protein (GFP) is what enables the worms to display fluorescence under the microscope. GFP by itself, however, has absolutely no link to F45D3.4, so how does GFP being active and lighting up the worm have anything to do with F45D3.4 being expressed? The simple answer is that a link is formed between GFP and F45D3.4, allowing for the expression of F34D3.4 to cause a trigger activating GFP, which in turn causes the worms to light up.
| 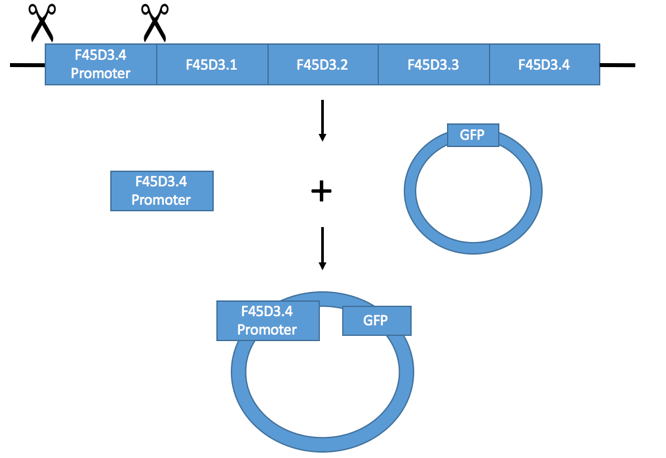
Figure 1: Illustration of F45D3.4 being incorporated into the pPD95.69 plasmid.
|
|
As seen in figure 1, the scissors represent the primers that are able to cut the promoter out and isolate it. Once the promoter is cloned, it can be integrated and placed into a plasmid (pPD95.69). A plasmid is a circular piece of DNA that is able to replicate independently from the host's chromosomal DNA [2]. The promoter can be integrated into the plasmid through enzymes that cut open the plasmid, and then paste in the gene in. The enzymes that facilitate this are called restriction enzymes. These enzymes are able to recognize a specific target sequence and then cut the DNA in two pieces (allowing for the gene to join). DNA ligase is then able to join the segments together, integrating the gene of interest into the plasmid [3]. As seen in the final product of figure 1, the plasmid has increased in size because the F45D3.4 promoter sequence is simply added in to the existing plasmid that contains GFP.
Once the plasmid contains the promoter of F45D3.4, it can be injected into the organism (process can be seen in figure 2). When injected into the roundworm, this plasmid is able to be integrated into the round worms genome (displayed by the bottom half of figure 2), and is now known as 3kb-F45D3.4::GFP. The two colons illustrate the pairing of F45D3.4 to GFP. As seen in the figure, by doing this, when protein X is present (representative of a transcription factor that binds in response to hypoxia) and is activated, it will bind to the promoter region, activating the promoter. As a result, GFP will also be activated. This activation is what produces the green fluorescence seen under the microscope. Alternatively, if protein X is not present, there is to bind to the promoter region. As a result, GFP is inactive and does not produce a green fluorescence. |
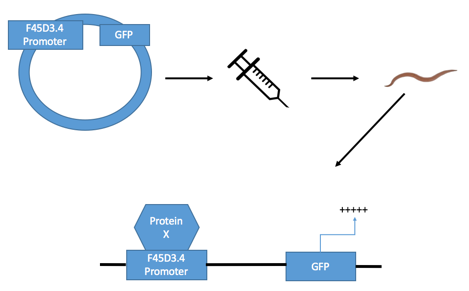
Figure 2: Illustration of the GFP construct (3kb-F45D3.4::GFP) being created.
|
References
References:
1. Huggett, J., & O'Grady, J. (2014, May). PCR Primers. Retrieved from https://www.highveld.com/pcr/pcr-primers.html 2. Kazilek. (2010, April 12). Plasmids. Retrieved from https://askabiologist.asu.edu/plasmids 3. Overview: DNA cloning. (n.d.). Retrieved from https://www.khanacademy.org/science/biology/biotech-dna-technology/dna-cloning-tutorial/a/overview-dna-cloning |