This experimental summary will be going over how the bacterial meta-interactome can be used to add to protein function prediction by increasing the amount of interactions known for a given protein. In this case, there were eight well-studied bacteria’s protein pool that were used to form this meta-interactome.
Wuchty et al. (2017)
The Protein Interactome of Streptococcus pneumoniae and
Bacterial Meta-interactomes Improve Function Predictions.
mSystems 2:1-10
The Protein Interactome of Streptococcus pneumoniae and
Bacterial Meta-interactomes Improve Function Predictions.
mSystems 2:1-10
(Translated by Karun Rajesh)
Experiment: Formation of Bacterial Meta-Interactome
|
Streptococcus pneumoniae is a pathogenic bacterium that can cause many types of illnesses, with many of these illnesses being life-threatening (bloodstream infections, pneumonia, meningitis, ear infections, etc.) These bacteria can invade the lungs, bloodstream, spinal cord, and tissues resulting in these life-threatening illnesses. It is vital to find more out about this bacterium in order to better treat and deal with it. Almost a third of the proteins’ functions in S. pneumoniae; however, are unknown. Knowledge of protein functions reveal the underlying characteristics of bacteria, allowing us to understand these bacteria more and possibly open up different avenues to deal with these pathogenic bacteria. In order to find out the unknown protein functions of S. pneumoniae, Wuchty et al. utilized a yeast-2-hybrid method approach alongside a bacterial meta-interactome with other well-studied bacteria to characterize these unknown protein functions. A yeast-2-hybrid approach is a common method of deriving protein function and interactions by taking two proteins and observing whether these proteins interact or not. A bacterial meta-interactome can be defined as all the molecular interactions between evolutionary conserved proteins in other bacteria.
|
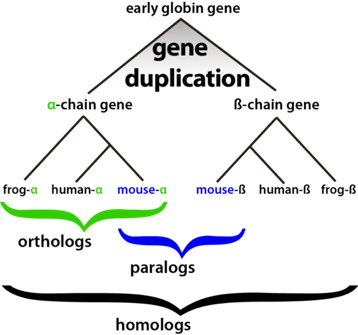
Fig. 3: Orthologous Proteins Example Orthologous proteins are proteins from different species that derived from the same ancestor. In this case, the orthologous proteins are the alpha proteins in the frog, mouse, human that derived from alpha chain gene.
|
|
In order to form the meta interactome, the protein orthologs in S. pneumoniae needed to be found in the other 8 bacteria. Genes in different species that evolved from a common ancestor. Orthologs usually retain the same function in the course of evolution. The program all-versus-all BLASTp was used to actually determine the orthologous groups between S. pneumoniae and these well-studied bacteria. BLASTp is a program used to compare primary biological sequence information, in this case being proteins. Given a set of proteins, BLASTp will return the most similar proteins from the protein database specified by the user. Protein sets of 2 species were run through this program and the sets with sequence pairs resulting in the best matching scores, score matching described in Figure 2, were classified as orthologous pairs. These groups characterized the orthologous proteins of S. pneumoniae and the set of 8 bacteria. The orthologous proteins found in the eight well-studied bacteria had their protein to protein interactions analyzed. These interactions from the orthologous proteins were then used as an add on to the yeast-2-hybrid interactions found for S. pneumoniae, which are protein interactions found within S. pnemoniae proteins. The set of these interactions with all the orthologous proteins is what makes up with the bacterial meta-interactome. |
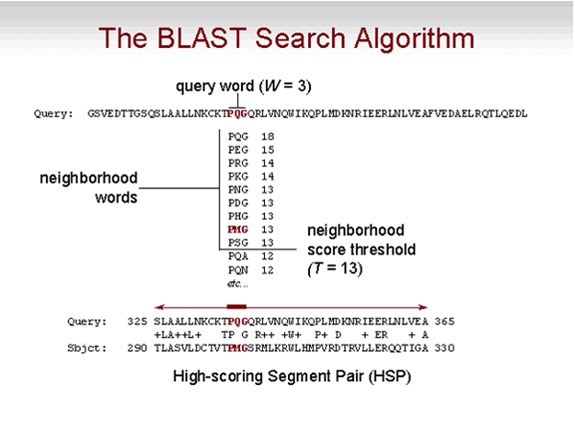
Fig. 4: BLASTp Algorithm BLASTp looks at a given protein sequence and checks whether a given set of amino acids in the sequence is similar/same as that of the protein sequence it is being compared to at the same position. This similarity is assigned a score and these scores are added to eventually produce the total score between the two protein sequences.
|