| The active site was the only location of PFOR made less rigid and only allowed to interact with the inhibitor. In a realistic setting, many other areas of PFOR may indirectly influence the activity of the active site. In another simulation experiment, more areas of the PFOR site should be made less rigid and the amixicile-PFOR interaction should be executed in order to observe any potential changes that could be traced to another area in PFOR. Furthermore, it would also be of interest to examine how would amixicile interact to a mutated active site of PFOR. It was predetermined that certain amino acid residues in the active site of PFOR (including ThrB31 and ArgB114, blue arrow) are conserved among anaerobic bacteria, from which it could inferred that amixicile (since it is predicted to bind to ThrB31 and ArgB114 of D. africanus) may be able to block pyruvate in a wide range of anaerobic bacteria. Since bacteria have the tendency to mutate and evolve at a fairly rapid rate, amixicile should be tested with possible mutants of PFOR to observe any potential changes in their interaction. |
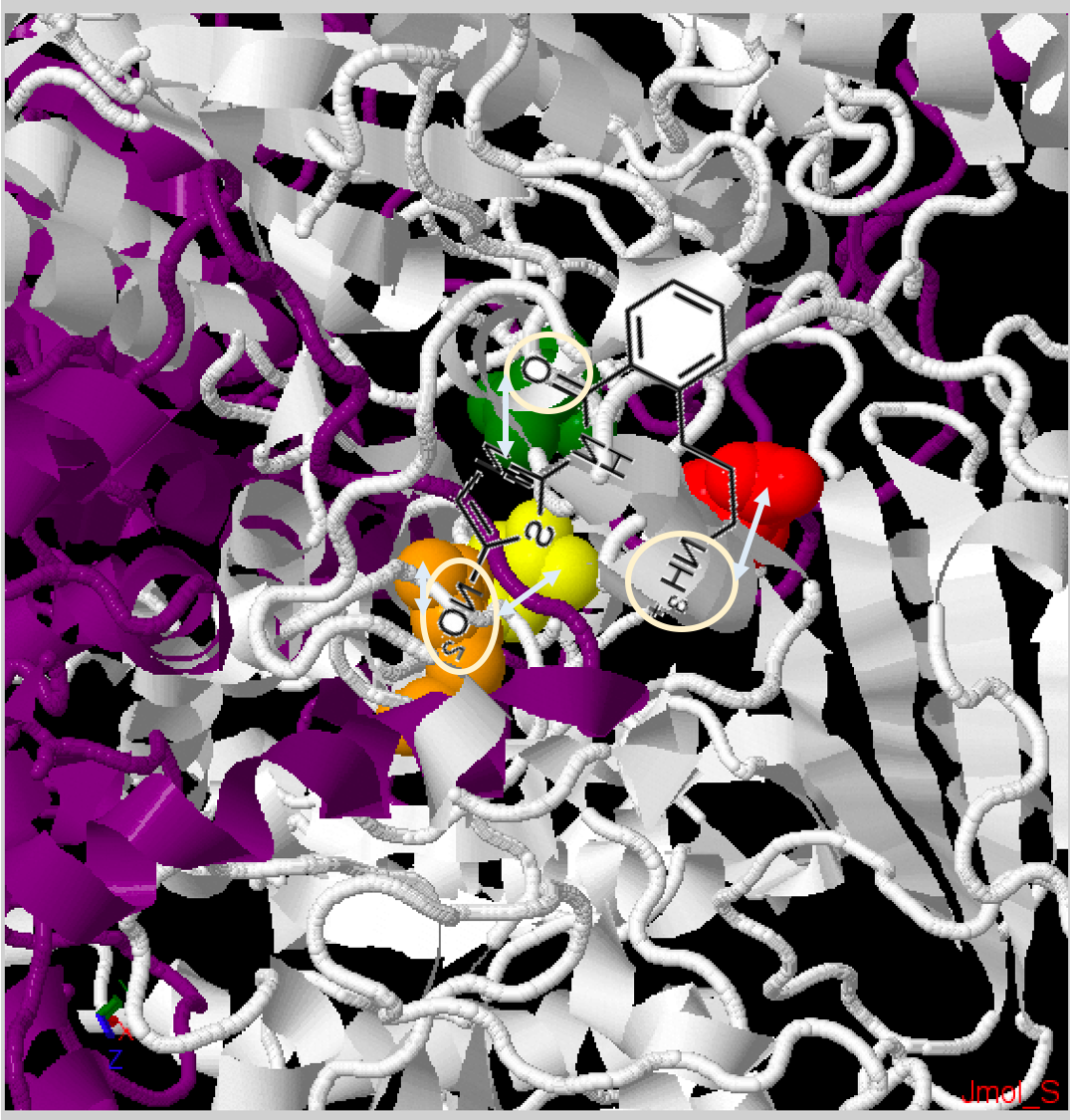
Fig. 3B: Zoomed in PFOR structure of Desulfovibrio africanus, particularly the active site. Chain A - Dark Purple, Chain B - White. Squared in white are the amino residues that the simulation predicts interact with amixicile: ThrB31 - yellow, ArgB114 - orange, AsnB996 - green, AspB456 - red. [Source: Jmol Protein Explorer].
|
Kennedy A.J., Bruce A.M., Gineste C., Ballard T.E., Olekhnovich I.N., Macdonald T.L. & Hoffman P.S. (July 2016)
Synthesis and Antimicrobial Evaluation of Amixicile-Based Inhibitors of the Pyruvate-Ferredoxin Oxidoreductases of Anaerobic Bacteria and Epsilonproteobacteria
Antimicrobial Agents and Chemotherapy 60:3980 - 3987
Synthesis and Antimicrobial Evaluation of Amixicile-Based Inhibitors of the Pyruvate-Ferredoxin Oxidoreductases of Anaerobic Bacteria and Epsilonproteobacteria
Antimicrobial Agents and Chemotherapy 60:3980 - 3987
(Translated by Nasita Islam)
PFOR structure
|
The pyruvate ferredoxin oxidoreductase (PFOR) crystal structure of D. africanus, an anaerobic bacteria was accessed from protein data bank (PDB ID: 1B0P). PDB is a library of proteins where each protein has a specific ID which allows for quick accession and observation of its crystal structure. Since previous studies reported observations of nitazoxanide (NTZ) binding with TPP in PFOR, acting as a competitive inhibitor to pyruvate by blocking pyruvate from bidning to TPP, therefore a non-reactive PFOR crystal structure with a bound pyruvate inspired the structure used in docking simulation. In other words, since NTZ was not predicted to change the structure of PFOR itself and rather act as a non-reactive barrier, an unresponsive PFOR crystal structure with a bound pyruvate was used in the MOE simulation.
|