| In figure 6, taking cLogP, PFOR inhibition percentage, MIC value all into consideration, amixicile analogue 4.5 appears to be promising for these reasons: it has the highest cLogP value, 2.81, indicating that it is the most hydrophobic, it also has the highest PFOR inhibition percentage, 95%, and has relatively low MIC values, 4 and 1 mm/mL for H. pylori and C. difficile respectively. It must be acknowledged however, that the enzyme (PFOR) inhibition percentage has very little correlation with overall biological (bacteria) inhibition. From this, it may be inferred that the amixicile analogue with the 4.5 R-group may impede PFOR’s activity in H. pylori and C. difficile significantly compared to the other analogues and amixicile but may vary in biological activity. |
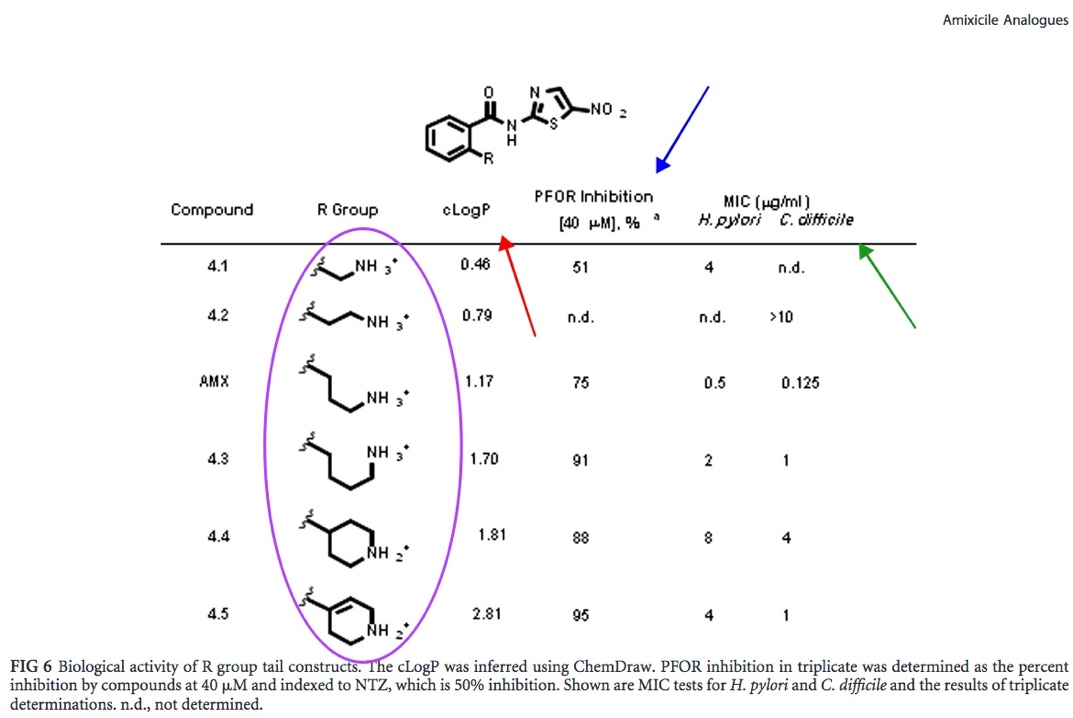
Fig. 6: Enzyme and MIC activity results [Kennedy et al.].
|
| In figure 7, while the highest percentage of PFOR inhibition is 97%, is attributed to compound 4.10 (an analogue of amixicile), the MIC values vary across the bacteria. The lowest relative MIC values can be attributed to amixicile analogue 4.8 with a fairly high PFOR inhibition percentage of 93%. Still, the enzymatic and biological activities for each compound are rather inconsistent. While amixicile and its analogues definitely affect enzyme and biological activity, each bacteria differs with regard to the dosage of amixicile required to interfere with biological activity. Nevertheless, constructing the amixicile analogues via docking simulation first helped to narrow down the number of compounds that can bind effectively with PFOR and block pyruvate from further oxidation and joining with Acetyl CoA, later destined to be tested in the wet lab through MIC and enzyme assays. |
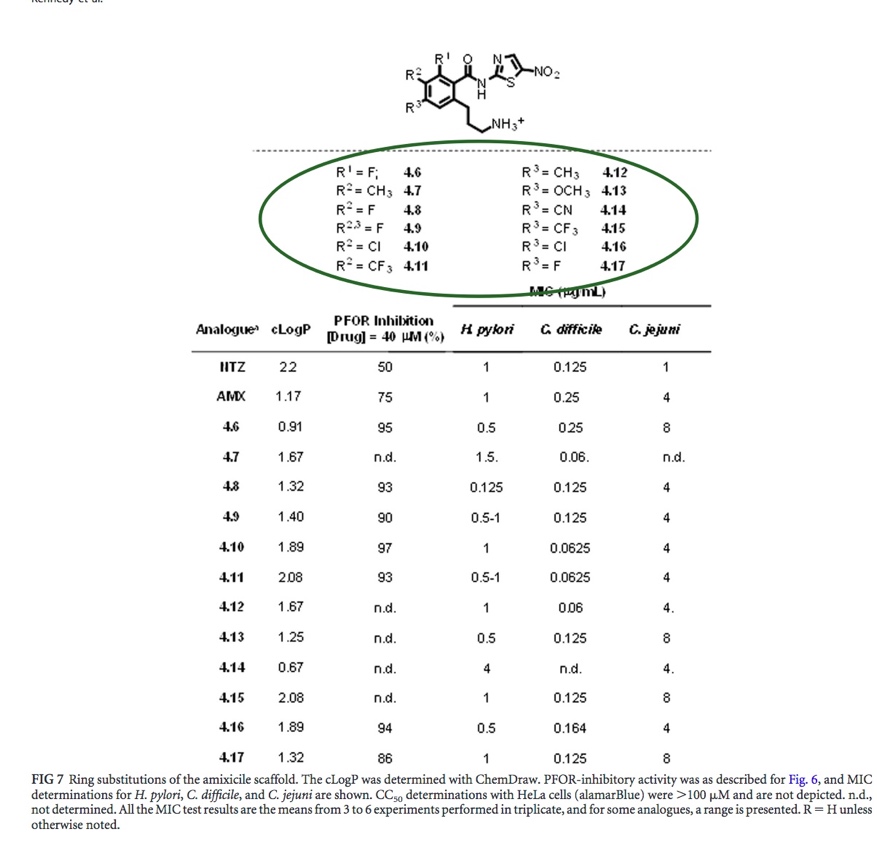
Fig. 7: Enzyme and MIC activity results [Kennedy et al.].
|