| Based on the observations of the docking simulation (where it was observed that the addition of a propylamine tail to amixicile projected increased binding affinity to the active site of PFOR), structure activity relationship (SAR) of different regions of amixicile was used to virtually generate and test various analogues of amixicile. SAR is a virtual method where the molecule in question is conceptually broken down and part(s) of the is/are analyzed and modified to observe its interaction with another molecule. As shown in figure 4, amixicile was divided into three regions (head, linker, and tail). The head region (propylamine tail) was modified and its projected interaction with specific molecules in the active site of PFOR was observed via MOE (molecular operating environment). In other words, variations of the propylamine were virtually constructed and the derived structures with the best [scores] would move on to the in vitro stage of this study (i.e. - the derived structures would be created by Suzuki coupling). The nitro group of the propylamine tail (circled in blue in Figure 4) and the tail (circled in purple in figure 4) was the main target of modification. |
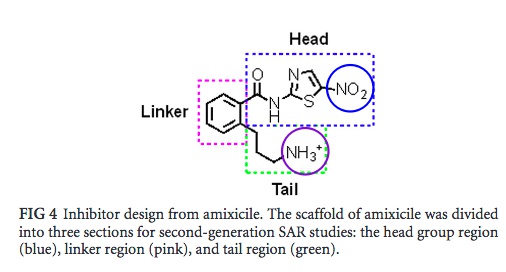
Fig. 4: Sectioned structure of amixicile. Each section was virtually manipulated to further optimize the efficiency of the amixicile analogues generated in the Docking Simulation experiment.
|
Motivation
Based on the amixicile analogues generated with the SAR method, the amixicile analogues with the highest scores were chemically crafted through Suzuki coupling. The chemically synthesized amixicile analogues are destined for the last experiment, minimum inhibitory concentration (MIC), where the amixcile analogues will be physically tested for its degree of inhibition (a clinical experiment). Procedure
|
Step 1: Synthesize actual propylamine tail (Suzuki coupling of propylamine tail)
The amine tails from the top-scoring amixicile analogues (Figure 5, blue circle) were separately synthesized first. Step 2: Add propylamine tail to X (Methyl Ester Saponification) The head group (Figure 5, green circle) was extended with either of the amine tails (blue circle) |
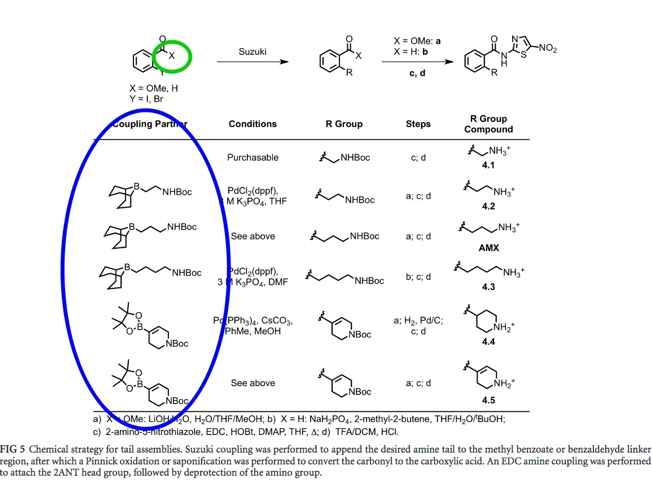
Fig. 5: Sectioned structure of amixicile. Each section was virtually manipulated to further optimize the efficiency of the amixicile analogues generated in the Docking Simulation experiment.
|
|
The R group was (Figure 5, yellow circle) was also subject to extension. As it was determined with the propylamine tail, the R group extensions (Figure 5, orange circle) were determined based on the top-scoring amixicile analogues.
Result The final synthesized amixicile analogues (Figure 5, sea-green circle) each have a different pair of propylamine tail (Extension of X group, blue circle) and head group (extension of R group, yellow circle). The compounds in the pink circle are similar to that of orange circle, except that they each have a distinguishable label (ex. 4.1, 4.4) |
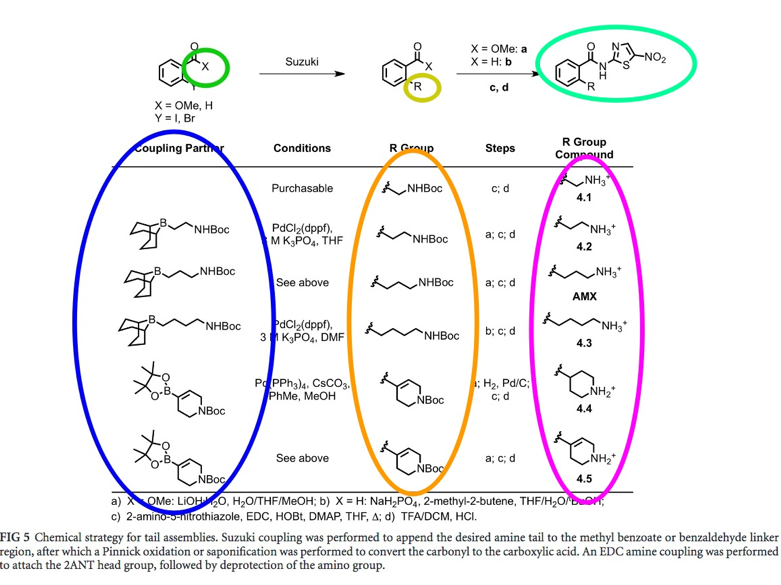
Fig. 5: Fig. 5: Molecular structure of amixicile analogues (determined by Docking Simulation). The tail group (circled in yellow) was subject to extended with different R groups (circled in orange) [Kennedy et al.]. The final analogue structures (scaffold circled sea-green) are circled in pink.
|