Enzyme assays and MIC (minimum inhibitory concentration) tests were both pharmacologically focused experiments. Both were implemented to observe the observe the rate of inhibition brought by each amixicile analogue (which was virtually and chemically synthesized via SAR and Suzuki coupling, respectively). The enzyme assay measured the percentage of inhibition of PFOR specifically while the MIC tests measured the percentage of inhibition of the bacteria overall. Procedure: Enzyme Assay
The PFOR of each bacterium (H. pylori and C. difficile) was amplified and isolated from E. coli. To prepare the PFOR for assay, potassium phosphate, sodium pyruvate, and benzyl viologen were added under anaerobic conditions - this procedure would prepare the PFOR for quantitative analysis so that inhibition can be measured via spectrophotometer. PFOR activity was initiated with or without an inhibitor (i.e. NTZ, amixicile, and the amixicile analogues) along with the addition of redox-active BV dye, which would act as an indicator of inhibition for the spectrophotometer, specifically any reduction activity would be highlighted by the BV dye.
Procedure: MIC Test
H. pylori and C. difficile were prepared differently for the MIC test, specifically via microdilution and agar dilution respectively.
Subprocedure: Agar dilution (C. difficile)
The bacteria were grown in a chopped-meat medium overnight, then transferred to fresh chopped-meat medium and let sit for 5 hours. The inhibitors (i.e. NTZ, amixicile, and the amixicile analogues) were separately diluted in agar media. Ten microliters of the bacteria-media mixture were then dispensed on each agar-inhibitor plate. The plates were incubated for 18 hours anaerobically and then observed with the naked eye for signs of growth.
Subprocedure: Microdilution (H. pylori)
The bacteria were grown in medium consisting of either brucella broth and brain heart. The cultures were diluted and placed in a 96-well microplate. The inhibitors (i.e. NTZ, amixicile, and the amixicile analogues) were serially diluted in the microplate. The microplate was then incubated and shaked at 37C aerobically. The turbidity in each well was then measured. Since turbidity usually indicates bacterial growth, the MIC value was calculated according to the concentration of the inhibitor in the well that had no detectable turbidity.
|
Result
The amixicile analogues (R groups of analogues circled in purple) were chemically synthesized via Suzuki coupling. The cLogP value (red arrow) determines the hydrophilicity of the specific R group - the higher the LogP value, the lower the hydrophilicity and therefore absorption. The PFOR inhibition value (blue arrow) attributes a percentage of inhibition of PFOR activity with each amixicile analogue (based on the spectrophotometer readings). Variations of the R group of the amixicile analogues were tested against the PFOR of H. pylori and C. difficile. It appears that the highest percentage of PFOR inhibition is 95%, is attributed to compound 4.5 (an analogue of amixicile). The MIC value is the minimum concentration of the inhibitor compound (in micrograms per microliters) where no turbidity was observed, presuming bacterium death. Note that the original amixicile structure (labeled AMX) has the lowest MIC values for both H. pylori and C. difficile, 0.5 and 0.125 ?m/mL respectively. Taking cLogP, PFOR inhibition percentage, MIC value all into consideration, amixicile analogue 4.5 appears to be promising for these reasons: it has the highest cLogP value, 2.81, indicating that it is the most hydrophobic, it also has the highest PFOR inhibition percentage, 95%, and has relatively low MIC values, 4 and 1 ?m/mL for H. pylori and C. difficile respectively. It must be acknowledged however, that the enzyme (PFOR) inhibition percentage has very little correlation with overall biological (bacteria) inhibition. |
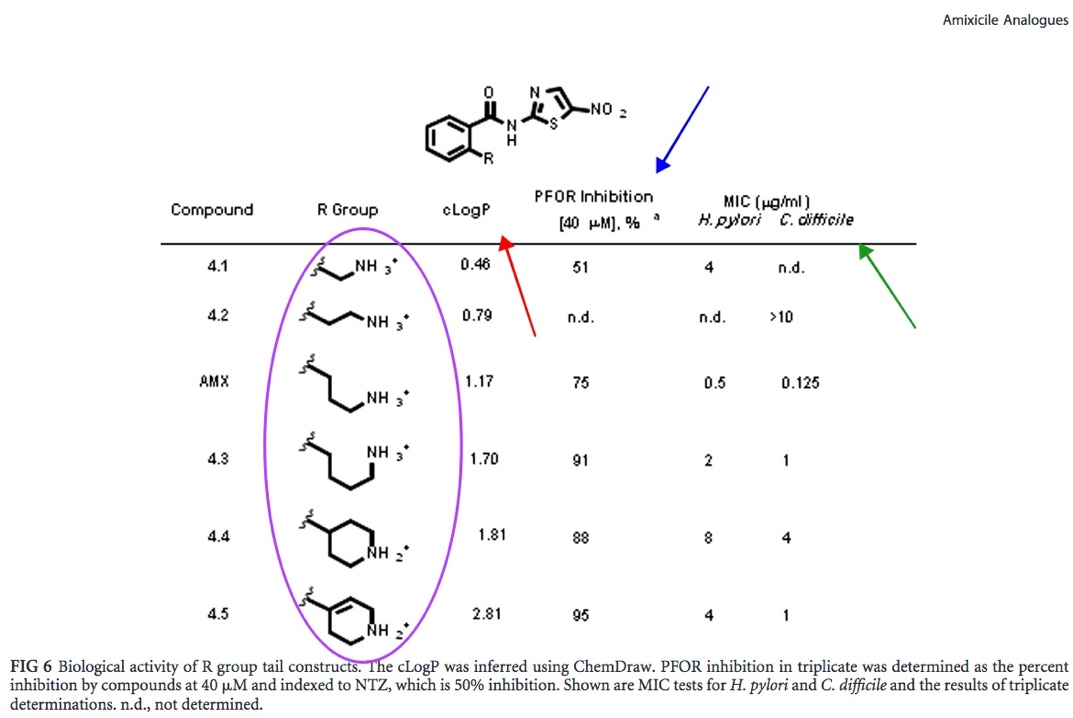
Fig. 6: Molecular structure of amixicile analogues (determined by Docking Simulation). The tail group (circled in yellow) was subject to extended with different R groups (circled in orange) [Kennedy et al.]. The final analogue structures (scaffold circled sea-green) are circled in pink.
|