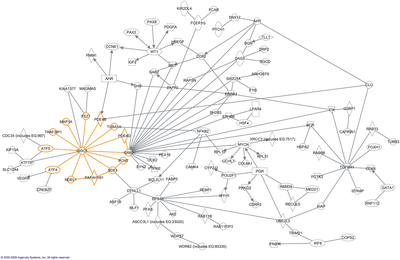
It is important to observe the protein networks of any given cell (whether bacterial or not) as these networks can show specifics on the different functions and relationships between proteins in a given cell. Examples of these functions include replication, translation, nucleotide transport, etc.
The image to the right is a graphic of an example protein network (also called an interactome) with genes represented by text in the boxes and interactions noted by the lines between the genes [1 Hennah W, Porteous D, (2009). The DISC1 pathway modulates expression of neurodevelopmental, synaptogenic and sensory perception genes. PLoS ONE 4(3) e4906.]. Graphics like these help many readers understand the relationships between proteins in a given network.
Little is known about the interactome of the human pathogen Streptococcus pneumoniae and to get a better understanding of its protein network would contribute to our ability to therapeutically intervene and ultimately find novel antibiotics to thwart any symptoms caused by this disease.
In general, many protein interactions are unknown in most species and model organisms, even when they are considered the backbone for numerous cellular activities. A theory of the majority rule
exists, where the functions of an unknown protein is hypothesized based on its interaction partners that are found in the protein network, also known as its interactome.