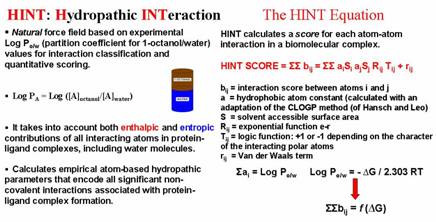
|
Research New Methods of Computer-Aided Molecular Design HINT. We have developed a unique new model, HINT (Hydropathic INTeractions), for understanding interactions in the biological environment. It is based on the classic solvent partitioning experiments. In the HINT model we use the experimental LogP information as the basis for studying interactions between molecules in the biological environment. The LogP parameter has significant experimental thermodynamic and interaction information encoded within it. The thermodynamic information is directly related to free energy, so that hydrophobicity includes entropy! The interaction information is a consequence of the fact that the underlying experiment is actually a measurement of interaction/environment preference -- between a polar, hydrogen-bonding solvent (water) and a hydrophobic solvent (octanol). These two environments and the non-covalent interactions they provide to the solute are not dissimilar from those that protein and other macromolecular receptors provide to bound ligands.
3D QSAR. We have been working on new algorithms for 3D QSAR. The first of these, LOCKSMITH, uses the HINT hydropathic field and simple statistics to describe the 3D hydropathic profile of a series of activity-related molecules. We are currently enhancing this model to make it predictive and to de novo design new molecules against the optimum 3D hydropathic structure. Recently we have been investigating algorithms for three-dimensional prediction of LogP utilizing hydropathic fields and/or and multipoles. Computational Titration. The titration algorithm allows the identification, modeling and optimization of the multiple protonation states of residues and ligand functional groups at the protein-ligand active site. This approach allows generating hypotheses on the best model for binding, i.e., the model with protonation corresponding to the optimal binding energy. The binding energy is evaluated with the HINT scoring function. In considering all the ionizable residues at the active site and modeling all the possible protonation states of residues and functional groups at this site, the computational titration algorithm represents fairly realistically the fluxional behavior of hydrogens in solvated biological systems. We are committed to making this tool available and are currently developing a web-enabled version that will be publicly accessible. de Novo Prediction Tools. One of the most promising, yet elusive goals of Computer- Aided Molecular Design (CAMD) is for the computer to design new, novel agents de novo, i.e., from the structure of the protein receptor/binding site. We are designing a new approach to this problem utilizing 3D hydropathic "key" maps of the receptor region. These maps encode significant structural information about the "ideal" ligand to bind at the receptor and will evaluate the spatial/hydropathic suitability of molecule fragments in putative active sites. Three-Dimensional Data Base Searching Algorithms. The most successful molecular modeling technique to date has been to search large corporate and commercial data bases of molecules of known three-dimensional structure with constrained structural parameter sets defining an "active" molecule or analog. With such a search, one can recover (and rediscover) molecules that may have unsuspected pharmacological activity. One of the major limitations of this approach is that only the most energetically favorable structure is likely to be stored in the database, and it is well known that bound ligands can adopt orientations and conformations that deviate significantly (in structure if not in energy) from the most favorable conformation. We have been looking at new data compression and analysis techniques that may make it possible to store more conformation/ energetics information about molecules in a three-dimensional database.
|