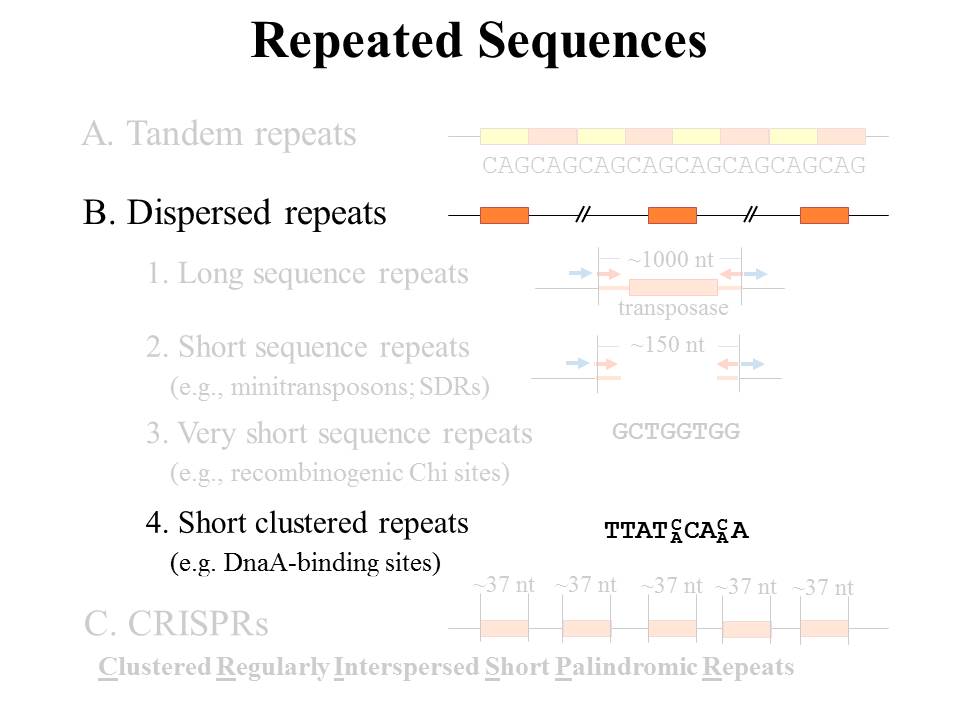
This category includes a variety of important short dispersed repeats, generally sequences that interact with proteins. We've seen these as clusters of very short sequences -- DnaA-binding sites and repressor-binding sites. In addition, you've encountered gapped palindromes that serve as a certain class of transcriptional terminator. Identifying these sites is certainly of interest, but greater insights can come from comparing them, either within a single genome or amongst several genomes. For example:
...and so forth.
- What other sequence features surround DnaA-binding sites from different organisms?
- What is the genetic context of such sites?
- Where are gapped palindromes found in certain phages?
- How distant are they from gene ends?
- How are common intergenic motifs preserved from one phage to the next?
Articles of possible interest:
- Petrillo M, Silvestro G, Paolo Di Nocera P, Boccia A, Paolella G (2006).
Stem-loop structures in prokaryotic genomes.
BMC Genomics 7:170
- Friedman DI, Court DL (2001).
Bacteriophage lambda: Alive and well and still doing its thing.
Curr Opin Microbiol 4:201-207.
- van Helden J, André B, Collado-Vides, J (1998).
Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies.
J Molec Biol 281:827-842.Notes of possible interest: