|
Where to get functions and strains for the simulation
|
If you have entered ViroBIKE and run the simulation file, then you
should have a BioBIKE screen in front of you containing a blue FUNCTIONS button at the top.
If you have not, then go to How to Get to the Simulation
and then come back here when you've gotten there.
If you mouse over the FUNCTIONS button, you'll see the special functions available
to you for the simulation (shown at right). To use a function, click its name on the FUNCTIONS menu,
and fill in the entry boxes in the function that comes down into your work space.
Similarly, you have a list of strains available to you, under the VARIABLES button.
|
|
Steps in doing the simulation
A. Obtain suppressor mutants
- Verify that the given mutant, FC0 (available from the VARIABLES button), is indeed R- and therefore unable to form plaques on E. coli K12. Use the CROSS function to test this, entering FC0 as both Strain 1 and Strain 1. Enter any number you like in the number box. Verify also that FC0 is able to form plaques on E. coli B.
- Use the MUTAGENIZE function
to obtain mutants derived from strain FC0 that are R+, i.e. able to form
plaques on E. coli K12. Almost all of these should be the result of suppressor mutations. Different R+ mutants might have different suppressor mutations, in different locations from one another. It will be useful to have a few suppressors.
- Use the DEFINE-STRAIN function to
grow up and name a few phage strains from the plaques of the previous step. Create a strain name that is unique to you, to make it easier to share without conflicting with the names others have created. For example, you might use your initials followed by a number. Keep a log of each strain and how it was produced! If you don't, the significance of strains will fade away from your mind, and the work to produce them will have been wasted.
- Use the CROSS function to
separate the suppressor mutations in your new strains from the mutation found in FC0.
You can do this by crossing each of your new strains against the wild-type strain, all now available
from the VARIABLES button. Both parent strains (your and wild-type) should be R+, therefore forming small plaques on E. coli B. Recombinant progeny should differ by having an R- phenotype, therefore forming large plaques on E. coli B. This type of cross should yield only two kinds of R- strains: those containing the FC0 (+) mutation and those containing the (-) suppressor mutation. Since you don't know which is which, you'll need to collect several plaques.
- Use the DEFINE-STRAIN function to
grow up and name several phage strains from the plaques of the previous step.
- Use the CROSS function to determine which
of the new recombinants are phage carrying the FC0 mutation (hence redundant) or the new mutation made in
Step A.1 (hence worth saving). You can do this by crossing the strains from Step 4 against FC0 to see if it is possible to
obtain wild-type (R+) recombinants, most easily detected by plating on E. coli K12. A cross between an FC0 recombinant and the original FC0 can not yield R+ recombinants on E. coli K12. Other recombinants shouldyield R+ recombinants in a cross with the original FC0.
B. Map the new mutations
Use the CROSS function to determine the frequency of recombination for each mutant crossed with a reference mutant (available from the VARIABLES button), e.g. FC0. Recombination frequency should be expressed in common units, the number of recombinant phages divided by the total number of progeny. Both quantities pose problems:
- The number of recombinant phages is not necessarily the number of plaques. In the example shown to the right, plating the cross on E. coli K12 will produce plaques only from recombinant wild-type phage, but progeny with both mutations (FC0 and H230) are also recombinants, even though they don't form plaques. Presume that they arise in equal number as wild-type and calculate the number of recombinant phages as double the number of plaques on E. coli K12. However, if the double mutant CAN form plaques on K12 (e.g. if the H230 mutation suppressed the FC0 mutation), then the number of recombinant phages is equal to the number of plaques on E. coli K12.
- The total number of progeny is definitely not the same as the number of phage participating in the cross. You can't directly measure the total number of progeny, since the great majority of the progeny won't form plaques on K12. Fortunately, you have a good surrogate measure -- plating on E. coli B.
|
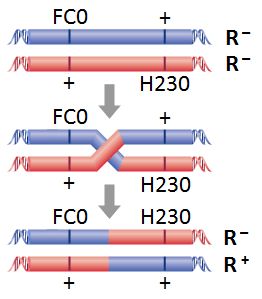
Figure 3: Recombination between FC0 and a hypothetical strain. (Thanks to Richard Twyman, U Warwick)
|
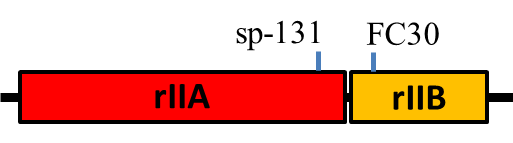
Mapping is more than a distance -- you need also to determine whether the new mutation sits to the left or right of FC0 (closer or more distant from rIIA on the genetic map below). Crossing your new mutant with one or more of the other given strains (FC30 and SP-131) should make this determination straight forward.
Accuracy is paramount. You should repeat the experiment to get an idea as to the variability of the map distances you calculate. You'll find that the variabiity is crushing with low numbers of plaques but quite tolerable with high numbers. Do the experiment so that the numbers are as high as possible.
The positions of the mutations in the reference mutants are shown
roughly below, and further information can be gained from Crick et al (1961) [3], Benzer (1961) [8], and Shinedling et al (1987) [9].
C. Construct mutants with multiple mutations
- Use the CROSS function to
recombine mutations of the same sign from mutants with different mutations of that sign. This is
somewhat tedious, because the two parental strains will be R-, and the strain you're looking for
may also be R-. You will need to test many plaques. To minimize the number you need to test, choose
parental mutants whose mutations are as far away from each other as possible.
- Use the DEFINE-STRAIN function to
grow up and name phage strains from the plaques of the previous step.
- Use the CROSS function to
test whether the new strain carries mutations from both phage parents. You can do this by performing
two crosses with each new strain: new strain vs parent 1 and new strain vs parent 2. If the new strain
carries both mutations, then both crosses should yield no wild-type (R+) recombinants when plated on
E. coli K12.
|


