BNFO301
– Introduction to Bioinformatics
Genome Analysis
|
A couple of weeks ago you had the idea
of studying Fragile X Syndrome in Drosophila.
The plan was to use the known human FMRP protein and then use it to scan
the Drosophila genome to find a similar gene. Then it's just a matter of
mutating the gene in Drosophila, and investigating what effect such a mutation
may have on fly mental function. Then you got stuck. No Drosophila genome means nothing to scan. So for the past two
weeks you have been sequencing the Drosophila
genome. Now that's behind you, and you have a genome sequence in hand.
The time has come to continue with the plan. 1. From the course web site, click on Resources and Links and
then on NCBI ( 2. Click on the down arrow to expand the choices for
the Search box from All Databases, and select Protein. 3. Enter FMRP into the for box, and press Go (or press Enter). 4. You might expect to pull up an entry for the human
FMRP gene, but no such luck. The search returned over 100 entries (!), some
from flies – which is interesting – some from mice... (This
number may be different depending on when you're doing this search). Let's
try to reduce the number to something more manageable. 5. Click on Preview/Index. At the bottom of the
resulting screen, you'll see a mechanism to add terms. Click on the down
arrow to expand the choices from All
fields and select Organism,
then type human into
the box to the right. Finally, click AND
(to specify that you're looking for entries that contain the word FMRP AND
the organism human), and click Go. 6. I got 24 entries now. Certainly an improvement, but
surely humans don't have 24 different FMRP proteins! The problem is that
we're searching multiple overlapping databases and getting the same protein
back multiple times. To cure that, click on Limits, expand the choices
in the right-most box from Only from, and select a single database (I
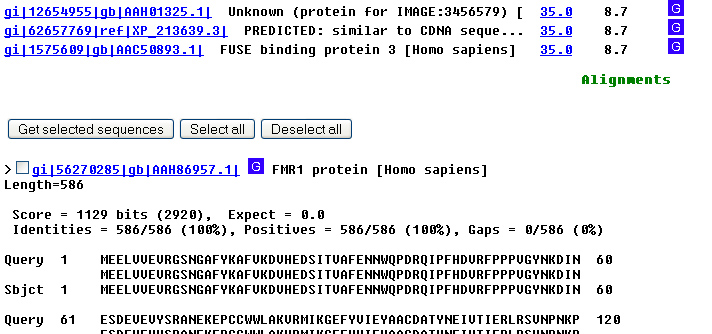
chose GenBank). Press Go again. 7. Now I'm down to 3 entries. They're different from
each other in ways that need not concern us now. Click on the entry with the
accession number AAH86957. 8. Lots of information here. One important item is a
reference to a journal article that describes the work that led to the
results you're looking at. Clicking on the PubMed number leads you to an
abstract and a link to the full length article. But what we're after right
now is the protein sequence, seen at the bottom of the page. The sequence is
given according to the one-letter amino acid codes (see course web site, Links and Resources,
Genetic Code, for a list of them). 9. The display is good for some purposes -- the
numbering makes it easy for humans to find what amino acid is at a specific
positions -- but computers prefer straight sequence, without numbers or
spaces. To get this, scroll back up to the top of the page, click on the down
arrow to expand the Display
box from GenPept, and select FastA. You probably already know what FastA
format is, but if you've gotten this far, you're now looking at it: One line
of documentation preceded by ">" and multiple lines of sequence.
If you want to save this file, click on the down arrow to expand the box
showing Send to, and select Text. Then use the browser to save
the file. Alternatively, just copy the sequence, with or without the
documentation line. 10.
Now we're ready
to do the search. Click on NCBI at
the top of the screen to return to NCBI home page, and click on Blast in the horizontal
toolbar near the top of the page. Blast (Basic Local Alignment Search Tool)
is undoubtedly the most widely used bioinformatic tool in existence. We'll
talk about what it does and how it does it, but for now, let's use it to find
the fruit-fly gene we want. We have a protein sequence, and we want to find a
fruit-fly protein, so look in the Protein box and click on
Protein-protein BLAST. 11.
Paste the FMRP
sequence into the Search
box and click on Blast.
You'll probably get the formatting BLAST page up pretty quickly,
showing conserved domains in the protein. That's for another day. For now,
click on Format to get the
search results. These results may take tens of seconds to come up or more or
less, depending on the time of day and alignment of the stars. 12.
Again, way too
many hits! We'll cut them down in a moment, but first,, scroll down until you
see things like:
This shows the
alignment of the protein we submitted (human FMR1 protein), the Query, to the
protein Blast found, the Subject. The best hit happens to be human FMR1
protein. No surprise! It found itself! Continue to scroll down and you'll see
similar proteins from other organisms: dog (Canis familiaris), cow (Bos
taurus), orangutan (Pongo pygmaeus), mouse, rat, chimp... the
amazing thing is that this protein is nearly identical amongst mammals. The
same is true of most protein. We share a common toolbox. 13.
Kill the
screen, returning to the formatting window. Scrolling down, you'll see that
you have the option of selecting which organism you want to focus on. Click
on the down arrow in the select from window, expanding the selection
from All organisms, and select Drosophila melanogaster[ORGN].
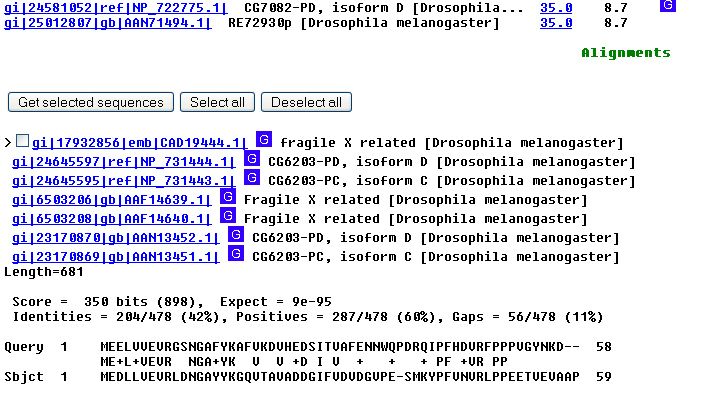
Then click Format again. 14.
Much better!
Now you have only a few hits, all from Drosophila. Here's the top hit: 15.
OK, we've got
the protein. What about the gene? We need that in order to
make a mutant fly that we can test for physiological function. Click on the GenBank
entry AAF14639.1. This gets you to the protein sequence. To find the gene
sequence, look at the DBSOURCE (database source) field. A protein sequence is
almost never obtained by direct sequencing of the protein but rather by
computer translation of the nucleotide sequence. The DBSOURCE is given as
AF205596.1. Click on that link. 16.
This brings you
to a page called "Drosophila melanogaster clone LD09557 Fragile X
related mRNA". The source of the protein sequence was virtual
translation of a cloned piece of mRNA (since you can't clone RNA directly, it
was first made into a DNA copy, or cDNA). Scroll down to the field called CDS
(CoDing Sequence -- don't ask me why anyone would think this is a proper
abbreviation!). It says that the coding sequence (some would say gene)
extends from position 423 to position 2468 in the cDNA sequence. Test that.
Scroll down to the cDNA sequence and find position 423. Do you find the
beginning of a start codon? Then find position 2468. Do you find the end of a
stop codon? 17.
This may seem a
roundabout path to get to the gene. Why not start with the human gene
and use it to pick out the fly gene? OK, let's do it that way.
Go back to step 2 and this time, instead of selecting Protein, select Nucleotide
(as before, confine the search to humans and to GenBank). This time even
after imposing limits, you still get dozens of sequences. 18.
Choose NM
002024, get the sequence from it as before and find your way to Blast
again. 19.
This time
choose Nucleotide-nucleotide Blast (blastn). Again paste the sequence into
the window and press Blast. Then press Format on the resulting
screen. When it comes back (probably a minute), you'll see that there are
tons of hits, but this time there are many mammalian sequences with multiple
differences with respect to the human sequence. But never mind that -- we're
after flies. So X out of the page, return to the format page, and limit the
output to Drosophila. 20.
What's this?
"No significant similarity found"??? Why
is that? How could it be that the protein search
gave five matches with extremely low expect values (definitely not merely by
chance) but the nucleotide search finds nothing at all? |
|
|