|
Goal of the experiment
Different theories about the genetic code -- overlapping vs non-overlapping, triplet vs quadruplet -- make very different predictions about how insertions and deletions of nucleotides within a gene will affect translation of that gene (see Question 2 of Problem Set 5). It follows that analyzing the phenotypes of different combinations of insertions and deletions ought to reveal a lot about the nature of the genetic code. In particular, three insertions or three deletions in close proximity figures to test the hypothesis that the code is non-overlapping triplet. Crick et al (1961) expended great effort to create such a mutant.
Prior art
Please see reference 3 for the original account of the experiment, references 4 and 5 for reviews of the principles behind the experiment, and references 2 and 6 for more detailed discussions of the experimental system used.
Key elements of the experimental system
The experiments were made possible using the remarkable characteristics of the infection by bacteriophage T4 of two strains of Escherichia coli, as discovered by Seymour Benzer [6]. In brief (and illustrated in Fig. 1):
- On E. coli strain B, wild-type bacteriophage T4 produces small plaques
- On E. coli strain B, derivatives of phage T4 defective in any r gene (rI, rIIA, or rIIB) produce large plaques.
- On E. coli strain K12, wild-type bacteriophage T4 forms plaques as on strain B.
- On E. coli strain K12, mutant phages defective in an r gene do not form plaques at all.
|
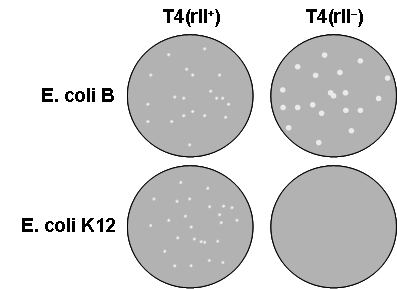
Figure 1: Infection of E. coli by phage T4. Ref. 5.
|
These characteristics make it possible to screen for mutant phages, looking for large plaques on strain B, and to select for wild-type recombinant phages, looking for plaques on strain K12.
|
A second key element of the experiment were the mutagens used to generate mutations. Mutagens typically alter DNA by changing one nucleotide to another, creating missense mutations. The mutagens employed by Crick et al (1961), acridine yellow and proflavine, instead created insertions or deletions by intercalating between the nucleotides during DNA replication [5,7], as illustrated in Fig. 2.
|
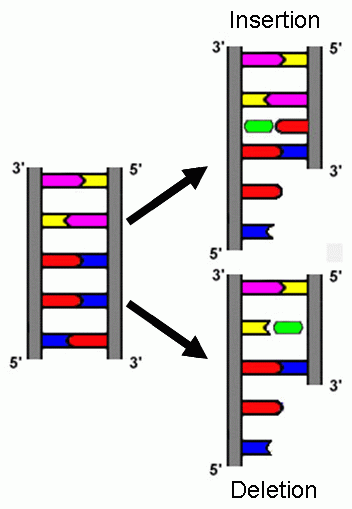
Figure 2: Insertion and deletion through acridine dye (green). Ref. 5.
|
Brief synopsis
- A proflavine-induced mutant (called
FC0) was obtained that was defective in r function through its ability to make large plaques in E. coli B. The underlying mutation was mapped to the rIIB gene of bacteriophage T4.
the first.
- Mutant FC0 was further mutagenized with
acridine yellow to produce mutants that have the wild-type ability to
produce infection on E. coli K12. These mutants carried two mutations:
the original (the same as in FC0) and a second mutation that suppressed
the first.
- The two mutations were separated, and FC0 was arbitrarily
labeled a (+) mutation and the suppressors (-) mutations, under the
theory that a (+) and a (-) mutation together could suppress each
other.
- By a similar means, (+) suppressors of the (-) suppressors
were obtained.
- Multiple (+) mutations were combined in a single bacteriophage to form strains with two and then three (+)
mutations.
|

