BIOL105
Proteins
& Their Basic Structure
Outline:
A. What can protein do?
DNA is often depicted as the blueprint of the cell. A
blueprint is something
an architect refers to in building a structure. It
contains a representation
of the final shape, its dimensions, whatís
connected to what, and
so forth. If you examine DNA, you will find none of
this. The molecule has
no knowledge of the cell's final shape, nor any
other of the things that
characterize blueprints. DNA merely lists the
components that make up the
proteins of a cell. But that is
enough.
The weight of action, then, lies
squarely on protein. Table 1 gives a synopsis of some
functions performed
by protein. At the top of the list is the catalysis of
the chemical reactions,
as emphasized in the last section. The enzyme
tyrosine hydroxylase, for
example, catalyzes the conversion of tyrosine to
the neurotransmitter L-DOPA.
Proteins are
responsible for other functions besides catalysis.
They are required for
the transport of a variety of compounds through membranes
or, in the case
of hemoglobin, the transport of oxygen in solution. Protein
also plays a
passive, structural role, for example in connective tissue.
There are many
other roles for protein, and Table 1 could have been many
times as big as
it is.
Table 1. Some
Biological Functions
of Proteins
FUNCTION |
EXAMPLE |
| Catalysis |
Tyrosine Hydroxylase (hormone & neurotransmitter
production)
| Binding:
transport |
Hemoglobin (oxygen
transport) |
| Binding:
defense |
Immunoglobins (immune
system) |
| Binding:
information |
Insulin (hormone) & Insulin
Receptor |
| Mechanical
Support |
Collagen (connective
tissue) |
| Mechanical
Work |
Actin/Myosin (muscle
contraction) |
B. What are proteins?
The function of a protein is determined ultimately by its
particular shape
and structure. At its most basic level, the structure of a
protein is simple.
It has to be, otherwise DNA could not specify it.
Understanding the structure
of protein thus answers two profound
questions:
How do proteins
control the activities of a cell?
How do
genes exert control over those
activities?
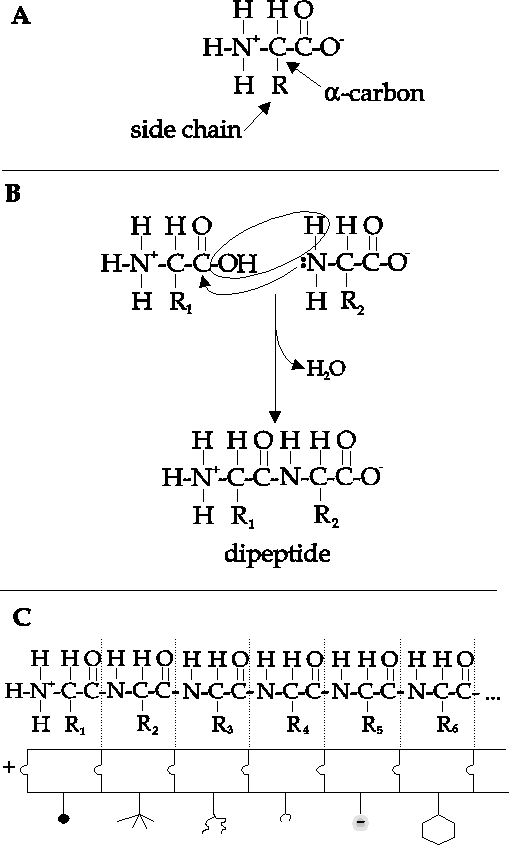 In brief, a protein is a linear array
of amino acids. If you grasp
all that sentence has to say, then you've come
a long way towards
understanding protein. Notice the pattern in Figure 1c.
A protein is a
polymer of a unit repeated again and again. That unit is
an amino acid.
Amino acids have some parts of their structure in common,
but they differ
from each other in one key position, the one labelled R
in the
diagram.
In brief, a protein is a linear array
of amino acids. If you grasp
all that sentence has to say, then you've come
a long way towards
understanding protein. Notice the pattern in Figure 1c.
A protein is a
polymer of a unit repeated again and again. That unit is
an amino acid.
Amino acids have some parts of their structure in common,
but they differ
from each other in one key position, the one labelled R
in the
diagram.
The synthesis of proteins is the
process of combining amino
acids in a linear chain. The backbone of this
chain is identical for all
proteins. If the R groups were similarly
invariable, then all proteins would
be alike, and protein would be able to
do only one thing, a not very interesting
thing at
that.
Figure 1.
Protein as a polymer
of alpha-amino acids. 1a. Structure of an amino
acid. "R"
represents side group, as shown in Figure 2. 1b.
Formation of dipeptide
by joining two amino acids. 1c. Polypeptide
chain composed of linked
amino acids. The shapes represent the different
R-groups, each with its
own chemical
properties.
Fortunately, the R
groups vary from one amino acid to the
next, amongst the 20 possibilities
shown in Figure
2. This listing of the twenty
major amino acids is a very good list
to get to know, but not to memorize.
If you go into biochemistry, you'll
find that they will become etched into
your brain without having to memorize
them, and if you don't, there's
probably no need to know the structures.
R
groups differ in their chemistry. Some are acidic under
normal conditions,
while others are basic or neutral. The charged amino
acids interact
strongly with water and so we call them hydrophilic. There
are other R
groups that interact strongly with water but are uncharged.
For example,
serine contains a hydroxyl group (an OH group), just like water
does, and
it's no surprise that serine is hydrophilic. There are also
hydrophobic
amino acids, like leucine, whose R-groups would tend to
segregate away from
water, because they interact less strongly with water
than water does with
itself.
There are many
other properties in which the twenty amino
acids differ from one another:
some are bulky, some small. And so forth.
Each amino acid represents a
different flavor, and the structure and properties
of a protein are defined
by the properties and order of its amino acids:
its primary
structure.
There are only twenty amino acids
used to synthesize proteins,
which limits what proteins are possible in
nature. How constricting is this
limitation? Consider the number of
possible dipeptides (two amino acids
joined together by a peptide bond).
There are 20 possible amino acids in
the first position and 20 possible
amino acids in the second position. That
makes 202 = 400 possible
dipeptides. Similarly, there are 203 = 8000 possible
tripeptides. Proteins
range in size from a smallish 100 amino acids to a
1000. The number of
possible proteins in nature is therefore
staggering!
C. Structure and basis for catalysis
Unfortunately, knowing merely that proteins are linear arrays
if (alpha-amino
acids doesn't tell us how they can have the varied
properties required of
proteins in a living cell. In particular, it doesn't
explain how proteins
can act as catalysts. For this we have to see the
protein in three dimensions.
The protein hexokinase (Figure 3), is the enzyme
that begins the
degradation of glucose in the liver. If you were to see
this molecule, the
first thing you might notice is that the enzyme has a
hole just the right
size for glucose to fit into. The binding of glucose
to the enzyme alters
the enzyme in such a way that glucose cannot escape
unless the enzyme again
changes shape. This normally occurs only after the
reaction catalyzed by
the enzyme is complete. So glucose goes in and glucose
6-phosphate goes
out.
The function of hexokinase is clearly
tied up in its shape.
How did the protein get to this shape? We now know
that the amino acids
may interact with their neighbors to form coils or
other structures. These
local interactions lead to what is called the second
ary structure of a protein.
In some cases structures
common to several proteins with similar
functions have been identified.
There are many such motifs known, and it
is sometimes possible to guess the
function of a protein simply by knowing
its primary
structure.
Amino acids may have more distant
interactions with one
another, giving rise to the tertiary structure of a
protein, the folding
of a polypeptide chain in three dimensions. For
example, the hydrophobic
amino acids would tend to be sequestered in the
middle of the protein, away
from water, just as the hydrophobic chains of
soap aggregate to minimize
contact with water. Charged and other
hydrophilic amino acids would tend
to lie outside the protein. You can see
this so some extent with hexokinase
(Figure
3).
It may be, however, that any way the
chain may twist, there
is no folding that can avoid patches of hydrophobic
amino acids from appearing
at the surface of the protein. What then? In
some cases, further aggregation
may occur between separate protein chains,
so that in the end, the completely
assembled protein consists of multiple
chains formed by the interaction
between them. Such proteins are said to
have quaternary structure. An example
of this is the protein hemoglobin,
the oxygen-carrying protein in blood.
It consists of four separate
polypeptide chains that interact with each
other. Separately, each subunit
can bind oxygen, due in part to the oxygen-binding
molecule, heme, which
fits into a hole created by the tertiary structure.
But the regulation of
oxygen binding, essential to the functioning of hemoglobin
in the body, is
apparent only when four subunits aggregate together.
The positions of specific amino acids determine not only
the shape
of the protein but also its capacity for catalysis (Figure
4). The folding of chymotrypsin, a digestive
enzyme that catalyzes the
hydrolysis (breakdown) of ingested protein in the
gut, creates a local region
of the enzyme called the active site. The
folding happens to place the 195th
amino acid in the chain, serine, near a
hole that has the shape of the amino
acid phenylalanine. When a
phenylalanine within a protein you eat finds
its way into the
phenylalanine-shaped hole of chymotrypsin, the amide bond
adjacent to
phenylalanine is positioned close enough to serine-195 that
a chemical
reaction takes place, breaking the amide bond. Once that occurs,
the broken
protein is released. The ability of chymotrypsin to do this depends
upon
the precise geometry of the active site. is dependent upon a
serine
occurring precisely at position number 195 and upon folding
occurring that
places serine in exactly the right position relative to the
protein being
digested.
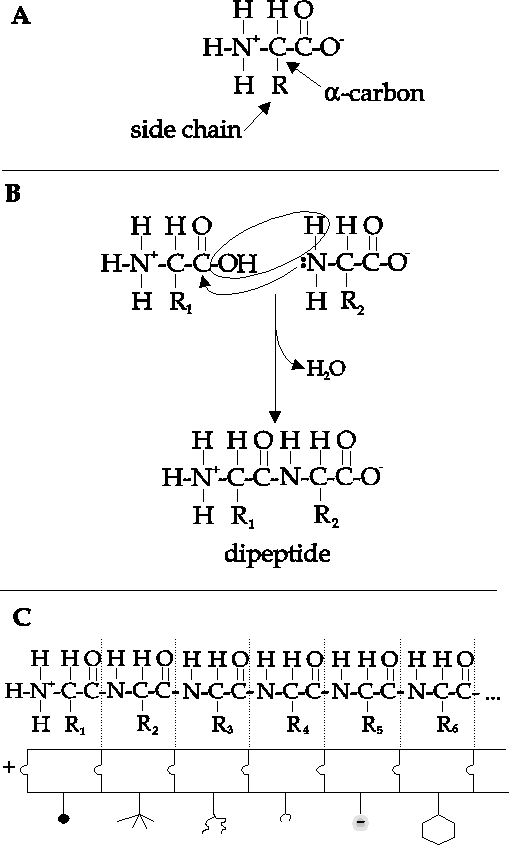 In brief, a protein is a linear array
of amino acids. If you grasp
all that sentence has to say, then you've come
a long way towards
understanding protein. Notice the pattern in Figure 1c.
A protein is a
polymer of a unit repeated again and again. That unit is
an amino acid.
Amino acids have some parts of their structure in common,
but they differ
from each other in one key position, the one labelled R
in the
diagram.
In brief, a protein is a linear array
of amino acids. If you grasp
all that sentence has to say, then you've come
a long way towards
understanding protein. Notice the pattern in Figure 1c.
A protein is a
polymer of a unit repeated again and again. That unit is
an amino acid.
Amino acids have some parts of their structure in common,
but they differ
from each other in one key position, the one labelled R
in the
diagram.